Miller Lab Research
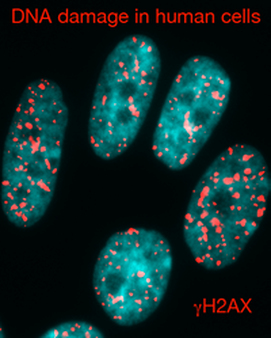
DNA damage represents a formidable challenge to genome maintenance and my lab studies the role of chromatin in these processes. To protect our genetic material, our cells have evolved multifaceted systems, collectively termed the DNA damage response (DDR), to detect and repair damaged DNA. It is clear that the true in vivo substrate of the DDR is not "naked" DNA but rather DNA assembled into chromatin. The structure and function of chromatin are regulated by histone modifications and chromatin modifying enzymes, which can markedly influence the DDR. Therefore, determining the interplay between the DDR and chromatin is fundamental for elucidating how cells maintain both epigenetic and genome integrity. The relevance of this research is highlighted by recent studies showing that mutations in many genes involved in the DDR and chromatin lead to cancer predisposition in humans. Therefore, we believe that deciphering the function of these pathways, both in normal and cancer cells, will contribute to the development of novel cancer therapies. Our research utilizes genetics, genomics, cell biology and molecular biology in both mouse and human tissue culture systems to gain insights into these areas of research. The lab also has interests in understanding anti-cancer drug mechanisms that function through DNA damage and chromatin. Many current drugs used in the clinic for cancer treatments act through DNA damage induction and pathways that regulate chromatin represent new targets for drug discovery. To explore this area of research, we employ a combination of chemical and molecular biology techniques to determine the interactions of small molecules (drugs) both at the cellular and molecular level. Taken together, the lab engages in an active research program that applies a multifaceted and diverse approach to these questions in hopes of defining the relationship between chromatin and the DDR, as well as gaining insights into the mechanisms of cancer therapeutic drugs that act at the chromatin and DNA level.
Ongoing projects in the lab include:
1. Role of Human Bromodomain Proteins in DNA double-strand break repair and genome stability
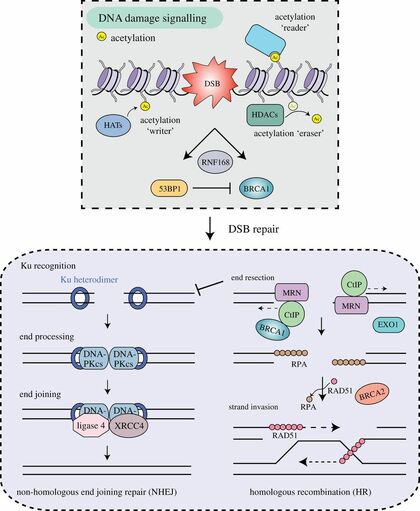
Chromatin-based DNA damage response (DDR) pathways are fundamental for preventing genome and epigenome instability, which are prevalent in cancer. An important chromatin DDR pathway is acetylation signaling which is controlled by histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes, which regulate acetylated lysines within proteins. Acetylated proteins, including histones, can modulate chromatin structure and provide molecular signals that are bound by acetyl-lysine binders, including bromodomain (BRD) proteins. Acetylation signaling regulates several DDR pathways, as exemplified by the preponderance of HATs, HDACs and BRD proteins that localize at DNA breaks to modify chromatin for lesion repair. We have elucidated the involvement of acetylation signaling in the DDR, focusing on the involvement of several BRD proteins in promoting chromatin remodeling to repair DNA double-strand breaks (Gong et al., Genes & Development, 2015; Gong et al., PLoS Genetics, 2016; Gong et al., Journal of Cell Biology, 2017; Gong and Miller, Cell Cycle, 2018). In cancer, DDR and acetylation pathways are often mutated or abnormally expressed. DNA damaging agents and drugs targeting epigenetic regulators, including HATs, HDACs, and BRD proteins, are used or are being developed to treat cancer. BRD proteins have widespread DDR functions including chromatin remodeling, chromatin modification and transcriptional regulation. Our research characterizes mechanistically how BRD proteins read acetylation signals within chromatin to trigger DDR and chromatin activities to facilitate genome–epigenome maintenance. In addition, we are working to define the relationship between BRD proteins and cancer, with an aim towards identifying therapeutic opportunities of these pathways in cancer.
2. Histone Variants and the DNA damage response
Genetic information is organized into chromatin, which consists of histones and proteins involved in regulating DNA compaction, accessibility and function. Chromatin is decorated by histone modifications, which provide signals that coordinate DNA-based processes including transcription and DNA damage response (DDR) pathways. Upon DNA damage, histone modifications are dynamically reshaped to accommodate DNA damage signaling and repair within chromatin. Chromatin connects DNA damage response factors to sites of damaged DNA to promote the signaling and repair of DNA lesions. The histone H2A variants H2AX, H2AZ, and macroH2A represent key chromatin constituents that facilitate DNA repair. Through proteomic screening of these variants, we have identified ZMYM3 (zinc finger, myeloproliferative, and mental retardation-type 3) as a chromatin-interacting protein that promotes DNA repair by homologous recombination (HR) (Leung et al., Genes and Development, 2017). Further research has recognized that ZMYM3 is a critical chromatin-binding DNA damage response factor, which modulates BRCA1 functions within chromatin to ensure the maintenance of genome integrity. In this project, we are working to better define the involvement of histone variants in the DDR. In addition, we are studying the emergent roles of ZnF domain-containing proteins in promoting genome integrity (Vilas et al., Trends in Genetics, 2018 ). Collectively, this work aims to understand how histone variants and proteins that interact with chromatin, including ZnF proteins, act to orchestrate genome integrity pathways and how these are involved in cancer.
3. Sources of Endogenous DNA damage – The DNA Damage-Up Proteins (i.e. DDPs)
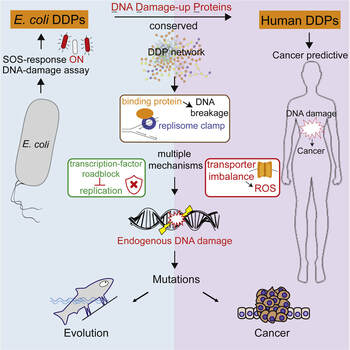
DNA damage provokes mutations and cancer and results from external carcinogens or endogenous cellular processes. However, the intrinsic instigators of endogenous DNA damage are poorly understood. We have identified proteins that promote endogenous DNA damage when overproduced: the DNA “damage-up” proteins (DDPs) (Xia et al., Cell, 2019). In a collaboration with Susan Rosenberg at Baylor College of Medicine, this project has identified a large network of DDPs in Escherichia coli and deconvoluted them into six function clusters, demonstrating DDP mechanisms in three: reactive oxygen increase by transmembrane transporters, chromosome loss by replisome binding, and replication stalling by transcription factors. Their 284 human homologs are over-represented among known cancer drivers, and their RNAs in tumors predict heavy mutagenesis and a poor prognosis. Half of the tested human homologs promote DNA damage and mutation when overproduced in human cells, with DNA damage-elevating mechanisms like those in E. coli. Our work identifies networks of DDPs that provoke endogenous DNA damage and may reveal DNA damage-associated functions of many human known and newly implicated cancer-promoting proteins. This project aims to understand and identify intrinsic protein mutagens (i.e. DDPs) involved in promoting endogenous DNA damage and mutations, which are known drivers of cancer initiation and progression.
Related publications:
For all publications, please click here
Gong F*, Chiu L-Y*, Cox B, Aymard F, Clouaire T, Leung JW, Cammarata M, Perez M, Agarwal P, Brodbelt JS, Legube G, Miller KM. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. (*authors contributed equally). Genes & Development (2015), 29:197-211. PMCID: PMC4298138 DOI: 10.1101/gad.252189.114
Gong, F., Couaire, T., Aguirrebengoa, M., Legube, G., and Miller KM. Histone demethylase KDM5A regulates ZMYND8-NuRD chromatin remodeler to promote DNA repair. Journal of Cell Biology. 2017. June 1st AOP. PMCID: PMC5496618 DOI: 10.1083/jcb.201611135
Spotlight: Price, B. KDM5A demethylase: Erasing histone modifications to promote repair of DNA breaks. Journal of Cell Biology. (2017). June 1st AOP.
Leung JW, Makharashvili N, Agarwal P, Chiu L-Y, Pourpre R, Cammarata MB, Cannon JR, Sherker A, Durocher D, Brodbelt JS, Paull TT and Miller KM. ZMYM3 regulates BRCA1 localization at damaged chromatin to promote DNA repair. Genes & Development, (2017). Feb 1;31(3):260-274. DOI: 10.1101/gad.292516.116
Xia J*, Chiu L-Y*, Nehring RB, Bravo Núñez MA, Mei Q, Perez M, Zhai Y, Fitzgerald DM, Pribis JP, Wang Y, Wang Y, Hu CW, Powell RT, LaBonte SA, Jalali A, Matadamas Guzmán ML, Lentzsch AM, Szafran AT, Joshi MC, Richters M, Gibson JL, Frisch RL, Hastings PJ, Bates D, Queitsch C, Hilsenbeck S, Coarfa C, Hu JC, Siegele DA, Scott KL, Liang H, Mancini MA, Herman C^, Miller KM^ and Rosenberg SM^. Bacteria-to-human protein networks reveal origins of endogenous DNA damage. Cell, (2019). (*Co-first author, ^co-corresponding authors). PMCID: PMC6344048 DOI: 10.1016/j.cell.2018.12.008
Research Highlights: Burgess, DJ. Human protein mutagens found via bacteria. Nature Reviews Genetics. (2019).
Kim, J.J.*, Lee, S.Y.*, Choi, J., Woo H.G., Xhemalce, B., Miller, K.M. (2020) PCAF-mediated histone acetylation promotes replication fork degradation by MRE11 and EXO1 in BRCA-deficient cells, Mol Cell. (*Co-first author) (published online September 22, 2020) DOI: 10.1016/j.molcel.2020.08.018
Highlighted in: Leuzzi G, Taglialatela A and Ciccia A. HATtracting Nucleases to Stalled Forks. Molecular Cell 80, Oct 15
Gong, F., Couaire, T., Aguirrebengoa, M., Legube, G., and Miller KM. Histone demethylase KDM5A regulates ZMYND8-NuRD chromatin remodeler to promote DNA repair. Journal of Cell Biology. 2017. June 1st AOP. PMCID: PMC5496618 DOI: 10.1083/jcb.201611135
Spotlight: Price, B. KDM5A demethylase: Erasing histone modifications to promote repair of DNA breaks. Journal of Cell Biology. (2017). June 1st AOP.
Leung JW, Makharashvili N, Agarwal P, Chiu L-Y, Pourpre R, Cammarata MB, Cannon JR, Sherker A, Durocher D, Brodbelt JS, Paull TT and Miller KM. ZMYM3 regulates BRCA1 localization at damaged chromatin to promote DNA repair. Genes & Development, (2017). Feb 1;31(3):260-274. DOI: 10.1101/gad.292516.116
Xia J*, Chiu L-Y*, Nehring RB, Bravo Núñez MA, Mei Q, Perez M, Zhai Y, Fitzgerald DM, Pribis JP, Wang Y, Wang Y, Hu CW, Powell RT, LaBonte SA, Jalali A, Matadamas Guzmán ML, Lentzsch AM, Szafran AT, Joshi MC, Richters M, Gibson JL, Frisch RL, Hastings PJ, Bates D, Queitsch C, Hilsenbeck S, Coarfa C, Hu JC, Siegele DA, Scott KL, Liang H, Mancini MA, Herman C^, Miller KM^ and Rosenberg SM^. Bacteria-to-human protein networks reveal origins of endogenous DNA damage. Cell, (2019). (*Co-first author, ^co-corresponding authors). PMCID: PMC6344048 DOI: 10.1016/j.cell.2018.12.008
Research Highlights: Burgess, DJ. Human protein mutagens found via bacteria. Nature Reviews Genetics. (2019).
Kim, J.J.*, Lee, S.Y.*, Choi, J., Woo H.G., Xhemalce, B., Miller, K.M. (2020) PCAF-mediated histone acetylation promotes replication fork degradation by MRE11 and EXO1 in BRCA-deficient cells, Mol Cell. (*Co-first author) (published online September 22, 2020) DOI: 10.1016/j.molcel.2020.08.018
Highlighted in: Leuzzi G, Taglialatela A and Ciccia A. HATtracting Nucleases to Stalled Forks. Molecular Cell 80, Oct 15